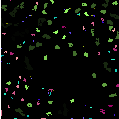In this chapter, the tools for multi-scale and multi-resoltuion processing (analysis, synthesis and fusion) will
be presented. Most of the algorithms are based on pyramidal approaches. These approaches
were first used for image compression and they are based on the fact that, once an image has
been low-pass filtered it does not have details beyond the cut-off frequency of the low-pass
filter any more. Therefore, the image can be subsampled – decimated – without any loss of
information.
A pyramidal decomposition is thus performed applying the following 3 steps in an iterative
way:
- Low pas filter the image In in order to produce F (In);
- Compute the difference Dn = In-F (In) which corresponds to the details at level n;
- Subsample F(In) in order to obtain In+1.
The result is a series of decrasing resolution images Ik and a series of decreasing resolution details
Dk.
If the smoothing filter used in the pyramidal analysis is a morphological filter, one cannot safely subsample
the filtered image without loss of information. However, by keeping the details possibly lost in the
down-sampling operation, such a decomposition can be used.
The Morphological Pyramid is an approach to such a decomposition. Its computation process is
an iterative analysis involving smoothing by the morphological filter, computing the details
lost in the smoothing, down-sampling the current image, and computing the details lost in the
down-sampling.
The source code for this example can be found in the file
Examples/MultiScale/MorphologicalPyramidAnalysisFilterExample.cxx.
This example illustrates the use of the otb::MorphologicalPyramidAnalyseFilter .
The first step required to use this filter is to include its header file.
#include "otbMorphologicalPyramidAnalysisFilter.h"
The mathematical morphology filters to be used have also to be included here.
#include "otbOpeningClosingMorphologicalFilter.h" #include "itkBinaryBallStructuringElement.h"
As usual, we start by defining the types needed for the pixels, the images, the image reader and the image
writer.
const unsigned int Dimension = 2; typedef unsigned char InputPixelType;
typedef unsigned char OutputPixelType; typedef otb::Image<InputPixelType, Dimension> InputImageType;
typedef otb::Image<OutputPixelType, Dimension> OutputImageType;
typedef otb::ImageFileReader<InputImageType> ReaderType;
typedef otb::ImageFileWriter<OutputImageType> WriterType;
Now, we define the types needed for the morphological filters which will be used to build the morphological
pyramid. The first thing to do is define the structuring element, which in our case, will be a
itk::BinaryBallStructuringElement which is templated over the pixel type and the dimension of the
image.
typedef itk::BinaryBallStructuringElement<InputPixelType, Dimension> StructuringElementType;
We can now define the type of the filter to be used by the morphological pyramid. In this case, we choose to
use an otb::OpeningClosingMorphologicalFilter which is just the concatenation of an opening and
a closing. This filter is templated over the input and output image types and the structuring element type that
we just define above.
typedef otb::OpeningClosingMorphologicalFilter<InputImageType, InputImageType,
StructuringElementType> OpeningClosingFilterType;
We can finally define the type of the morpholoical pyramid filter. The filter is templated over the input and
output image types and the lowpas morphological filter to be used.
typedef otb::MorphologicalPyramidAnalysisFilter<InputImageType, OutputImageType,
OpeningClosingFilterType> PyramidFilterType;
Since the otb::MorphologicalPyramidAnalyseFilter generates a list of images as output, it is useful
to have an iterator to access the images. This is done as follows :
typedef PyramidFilterType::OutputImageListType::Iterator ImageListIterator;
We can now instantiate the reader in order to access the input image which has to be analysed.
ReaderType::Pointer reader = ReaderType::New(); reader->SetFileName(inputFilename);
We instantiate the morphological pyramid analysis filter and set its parameters which are:
- the number of iterations or levels of the pyramid;
- the subsample scale or decimation factor between two successive pyramid levels.
After that, we plug the pipeline and run it by calling the Update() method.
PyramidFilterType::Pointer pyramid = PyramidFilterType::New(); pyramid->SetNumberOfLevels(numberOfLevels);
pyramid->SetDecimationRatio(decimationRatio); pyramid->SetInput(reader->GetOutput()); pyramid->Update();
The morphological pyramid has 5 types of output:
- the analysed image at each level of the pyramid through the GetOutput() method;
- the brighter details extracted from the filtering operation through the GetSupFilter()
method;
- the darker details extracted from the filtering operation through the GetInfFilter() method;
- the brighter details extracted from the resampling operation through the GetSupDeci()
method;
- the darker details extracted from the resampling operation through the GetInfDeci() method;
to decimation
Each one of these methods provides a list of images (one for each level of analysis), so we can iterate
through the image lists by using iterators.
ImageListIterator itAnalyse = pyramid->GetOutput()->Begin();
ImageListIterator itSupFilter = pyramid->GetSupFilter()->Begin();
ImageListIterator itInfFilter = pyramid->GetInfFilter()->Begin();
ImageListIterator itInfDeci = pyramid->GetSupDeci()->Begin();
ImageListIterator itSupDeci = pyramid->GetInfDeci()->Begin();
We can now instantiate a writer and use it to write all the images to files.
WriterType::Pointer writer = WriterType::New(); int i = 1; // Writing the results images
std::cout << (itAnalyse != (pyramid->GetOutput()->End())) << std::endl;
while (itAnalyse != pyramid->GetOutput()->End()) { writer->SetInput(itAnalyse.Get());
writer->SetFileName(argv[0 ⋆ 4 + i + 1]); writer->Update(); writer->SetInput(itSupFilter.Get());
writer->SetFileName(argv[1 ⋆ 4 + i + 1]); writer->Update(); writer->SetInput(itInfFilter.Get());
writer->SetFileName(argv[2 ⋆ 4 + i + 1]); writer->Update(); writer->SetInput(itInfDeci.Get());
writer->SetFileName(argv[3 ⋆ 4 + i + 1]); writer->Update(); writer->SetInput(itSupDeci.Get());
writer->SetFileName(argv[4 ⋆ 4 + i + 1]); writer->Update(); ++itAnalyse;
++itSupFilter; ++itInfFilter; ++itInfDeci; ++itSupDeci; ++i; }
Figure 15.1 shows the test image to be processed by the morphological pyramid.
Figure 15.2 shows the 4 levels of analysis of the image.
Figure 15.3 shows the 4 levels of bright details.
Figure 15.4 shows the 4 levels of dark details.
Figure 15.5 shows the 4 levels of bright decimation details.
Figure 15.6 shows the 4 levels of dark decimation details.
The source code for this example can be found in the file
Examples/MultiScale/MorphologicalPyramidSynthesisFilterExample.cxx.
This example illustrates the use of the otb::MorphologicalPyramidSynthesisFilter
.
The first step required to use this filter is to include its header file.
#include "otbMorphologicalPyramidSynthesisFilter.h"
The mathematical morphology filters to be used have also to be included here, as well as the
otb::MorphologicalPyramidAnalyseFilter in order to perform the analysis step.
#include "otbMorphologicalPyramidAnalysisFilter.h"
#include "otbOpeningClosingMorphologicalFilter.h" #include "itkBinaryBallStructuringElement.h"
As usual, we start by defining the types needed for the pixels, the images, the image reader and the image
writer.
const unsigned int Dimension = 2; typedef unsigned char InputPixelType;
typedef unsigned char OutputPixelType; typedef otb::Image<InputPixelType, Dimension> InputImageType;
typedef otb::Image<OutputPixelType, Dimension> OutputImageType;
typedef otb::ImageFileReader<InputImageType> ReaderType;
typedef otb::ImageFileWriter<OutputImageType> WriterType;
Now, we define the types needed for the morphological filters which will be used to build the morphological
pyramid. The first thing to do is define the structuring element, which in our case, will be a
itk::BinaryBallStructuringElement which is templated over the pixel type and the dimension of the
image.
typedef itk::BinaryBallStructuringElement<InputPixelType, Dimension> StructuringElementType;
We can now define the type of the filter to be used by the morphological pyramid. In this case, we choose to
use an otb::OpeningClosingMorphologicalFilter which is just the concatenation of an opening and
a closing. This filter is theplated over the input and output image types and the structurung element type that
we just define above.
typedef otb::OpeningClosingMorphologicalFilter<InputImageType, InputImageType,
StructuringElementType> OpeningClosingFilterType;
We can now define the type of the morpholoical pyramid filter. The filter is templated over the input and
output mage types and the lowpas morphological filter to be used.
typedef otb::MorphologicalPyramidAnalysisFilter<InputImageType, OutputImageType,
OpeningClosingFilterType> PyramidAnalysisFilterType;
We can finally define the type of the morpholoical pyramid synthesis filter. The filter is templated over the
input and output mage types.
typedef otb::MorphologicalPyramidSynthesisFilter<InputImageType, OutputImageType>
PyramidSynthesisFilterType;
We can now instantiate the reader in order to access the input image which has to be analysed.
ReaderType::Pointer reader = ReaderType::New(); reader->SetFileName(inputFilename);
We instantiate the morphological pyramid analysis filter and set its parameters which are:
- the number of iterations or levels of the pyramid;
- the subsample scale or decimation factor between two successive pyramid levels.
After that, we plug the pipeline and run it by calling the Update() method.
PyramidAnalysisFilterType::Pointer pyramidAnalysis = PyramidAnalysisFilterType::New();
pyramidAnalysis->SetNumberOfLevels(numberOfLevels); pyramidAnalysis->SetDecimationRatio(decimationRatio);
pyramidAnalysis->SetInput(reader->GetOutput()); pyramidAnalysis->Update();
Once the analysis step is finished we can proceed to the synthesis of the image from its different levels of
decomposition. The morphological pyramid has 5 types of output:
- the Analysisd image at each level of the pyramid through the GetOutput() method;
- the brighter details extracted from the filtering operation through the GetSupFilter()
method;
- the darker details extracted from the filtering operation through the GetInfFilter() method;
- the brighter details extracted from the resampling operation through the GetSupDeci()
method;
- the darker details extracted from the resampling operation through the GetInfDeci() method;
to decimation
This outputs can be used as input of the synthesis filter by using the appropriate methods.
PyramidSynthesisFilterType::Pointer pyramidSynthesis = PyramidSynthesisFilterType::New();
pyramidSynthesis->SetInput(pyramidAnalysis->GetOutput()->Back());
pyramidSynthesis->SetSupFilter(pyramidAnalysis->GetSupFilter());
pyramidSynthesis->SetSupDeci(pyramidAnalysis->GetSupDeci());
pyramidSynthesis->SetInfFilter(pyramidAnalysis->GetInfFilter());
pyramidSynthesis->SetInfDeci(pyramidAnalysis->GetInfDeci());
After that, we plug the pipeline and run it by calling the Update() method.
pyramidSynthesis->Update();
We finally instatiate a the writer in order to save the result image to a file.
WriterType::Pointer writer = WriterType::New(); writer->SetFileName(outputFilename);
writer->SetInput(pyramidSynthesis->GetOutput()->Back()); writer->Update();
Since the synthesis operation is applied on the result of the analysis, the input and the output images should
be identical. This is the case as shown in figure 15.7.
Of course, in a real application, a specific processing will be applied after the analysis and
before the synthesis to, for instance, denoise the image by removing pixels at the finer scales,
etc.
One of the possible uses of the morphological pyramid is the segmentation of objects – regions – of a
particular scale.
The source code for this example can be found in the file
Examples/MultiScale/MorphologicalPyramidSegmenterExample.cxx.
This example illustrates the use of the otb::MorphologicalPyramid::Segmenter . This class performs
the segmentation of a detail image extracted from a morphological pyramid analysis. The Segmentation is
performed using the itk::ConnectedThresholdImageFilter . The seeds are extracted from the image
using the otb::ImageToPointSetFilter . The thresolds are set by using quantiles computed with the
HistogramGenerator.
The first step required to use this filter is to include its header file.
#include "otbMorphologicalPyramidSegmenter.h"
As usual, we start by defining the types needed for the pixels, the images, the image reader and the image
writer. Note that, for this example, an RGB image will be created to store the results of the
segmentation.
const unsigned int Dimension = 2; typedef double InputPixelType;
typedef unsigned short LabelPixelType; typedef itk::RGBPixel<unsigned char> RGBPixelType;
typedef otb::Image<InputPixelType, Dimension> InputImageType;
typedef otb::Image<LabelPixelType, Dimension> LabelImageType;
typedef otb::Image<RGBPixelType, 2> RGBImageType;
typedef otb::ImageFileReader<InputImageType> ReaderType;
typedef otb::ImageFileWriter<RGBImageType> WriterType;
We define now the segmenter. Please pay attention to the fact that this class belongs to the
morphologicalPyramid namespace.
typedef otb::MorphologicalPyramid::Segmenter<InputImageType, LabelImageType> SegmenterType;
We instantiate the readers which will give us access to the image of details produced by the morphological
pyramid analysis and the original image (before analysis) which is used in order to produce
segmented regions which are sharper than what would have been obtained with the detail image
only.
ReaderType::Pointer reader = ReaderType::New(); reader->SetFileName(inputFilename);
ReaderType::Pointer reader2 = ReaderType::New(); reader2->SetFileName(originalFilename);
We instantiate the segmenter and set its parameters as follows. We plug the output of the readers for the
details image and the original image; we set the boolean variable which controls whether the segmented
details are bright or dark; we set the quantile used to threshold the details image in order to obtain the seed
points for the segmentation; we set the quantile for setting the threshold for the region growing
segmentation; and finally, we set the minimum size for a segmented region to be kept in the final
result.
SegmenterType::Pointer segmenter = SegmenterType::New(); segmenter->SetDetailsImage(reader->GetOutput());
segmenter->SetOriginalImage(reader2->GetOutput()); segmenter->SetSegmentDarkDetailsBool(segmentDark);
segmenter->SetSeedsQuantile(seedsQuantile); segmenter->SetConnectedThresholdQuantile(segmentationQuantile);
segmenter->SetMinimumObjectSize(minObjectSize);
The output of the segmenter is an image of integer labels, where a label denotes membership of a pixel in a
particular segmented region. This value is usually coded using 16 bits. This format is not practical for
visualization, so for the purposes of this example, we will convert it to RGB pixels. RGB images have the
advantage that they can be saved as a simple png file and viewed using any standard image
viewer software. The itk::Functor::ScalarToRGBPixelFunctor class is a special function
object designed to hash a scalar value into an itk::RGBPixel . Plugging this functor into the
itk::UnaryFunctorImageFilter creates an image filter for that converts scalar images to RGB
images.
typedef itk::Functor::ScalarToRGBPixelFunctor<LabelPixelType> ColorMapFunctorType;
typedef itk::UnaryFunctorImageFilter<LabelImageType, RGBImageType,
ColorMapFunctorType> ColorMapFilterType; ColorMapFilterType::Pointer colormapper = ColorMapFilterType::New();
We can now plug the final segment of the pipeline by using the color mapper and the image file
writer.
colormapper->SetInput(segmenter->GetOutput()); WriterType::Pointer writer = WriterType::New();
writer->SetInput(colormapper->GetOutput()); writer->SetFileName(outputFilename1); writer->Update();
Figure 15.8 shows the results of the segmentation of the image of bright details obtained with the
morphological pyramid analysis.
This same approach can be applied to all the levels of the morphological pyramid analysis.
The source code for this example can be found in the file
Examples/MultiScale/MorphologicalPyramidSegmentationExample.cxx.
This example illustrates the use of the otb::MorphologicalSegmentationFilter . This
filter performs a segmentation of the details supFilter and infFilter extracted with the
morphological pyramid. The segmentation algorithm used is based on seeds extraction using the
otb::ImageToPointSetFilter , followed by a connected threshold segmentation using the
itk::ConnectedThresholdImageFilter . The threshold for seeds extraction and segmentation are
computed using quantiles. A pre processing step is applied by multiplying the full resolution brighter details
(resp. darker details) with the original image (resp. the inverted original image). This performs an
enhancement of the regions contour precision. The details from the pyramid are set via the
SetBrighterDetails() and SetDarkerDetails() methods. The brighter and darker details depend on
the filter used in the pyramid analysis. If the otb::OpeningClosingMorphologicalFilter
filter is used, then the brighter details are those from the supFilter image list, whereas if the
otb::ClosingOpeningMorphologicalFilter filter is used, the brighter details are those from the
infFilter list. The output of the segmentation filter is a single segmentation images list, containing first
the brighter details segmentation from higher scale to lower, and then the darker details in the same order.
The attention of the user is drawn to the fact that since the label filter used internally will deal with a
large number of labels, the OutputPixelType is required to be sufficiently precise. Unsigned
short or Unsigned long would be a good choice, unless the user has a very good reason to think
that a less precise type will be sufficient. The first step to use this filter is to include its header
file.
#include "otbMorphologicalPyramidSegmentationFilter.h"
The mathematical morphology filters to be used have also to be included here, as well as the morphological
pyramid analysis filter.
#include "otbOpeningClosingMorphologicalFilter.h" #include "itkBinaryBallStructuringElement.h"
#include "otbMorphologicalPyramidAnalysisFilter.h"
As usual, we start by defining the types for the pixels, the images, the reader and the writer. We also define
the types needed for the morphological pyramid analysis.
const unsigned int Dimension = 2; typedef unsigned char InputPixelType;
typedef unsigned short OutputPixelType; typedef otb::Image<InputPixelType, Dimension> InputImageType;
typedef otb::Image<OutputPixelType, Dimension> OutputImageType;
typedef otb::ImageFileReader<InputImageType> ReaderType;
typedef otb::ImageFileWriter<OutputImageType> WriterType;
typedef itk::BinaryBallStructuringElement<InputPixelType, Dimension>
StructuringElementType; typedef otb::OpeningClosingMorphologicalFilter<InputImageType,
InputImageType, StructuringElementType> OpeningClosingFilterType;
typedef otb::MorphologicalPyramidAnalysisFilter<InputImageType, InputImageType,
OpeningClosingFilterType> PyramidFilterType;
We can now define the type for the otb::MorphologicalPyramidSegmentationFilter which is
templated over the input and output image types.
typedef otb::MorphologicalPyramidSegmentationFilter<InputImageType, OutputImageType>
SegmentationFilterType;
Since the output of the segmentation filter is a list of images, we define an iterator type which will be used
to access the segmented images.
typedef SegmentationFilterType::OutputImageListIteratorType OutputListIteratorType;
The following code snippet shows how to read the input image and perform the morphological pyramid
analysis.
ReaderType::Pointer reader = ReaderType::New(); reader->SetFileName(inputFilename);
PyramidFilterType::Pointer pyramid = PyramidFilterType::New(); pyramid->SetNumberOfLevels(numberOfLevels);
pyramid->SetDecimationRatio(decimationRatio); pyramid->SetInput(reader->GetOutput());
We can now instantiate the segmentation filter and set its parameters. As one can see, the
SetReferenceImage() is used to pass the original image in order to obtain sharp region boundaries. Using
the SetBrighterDetails() and SetDarkerDetails() the output of the analysis is passed to the
filter. Finally, the parameters for the segmentation are set by using the SetSeedsQuantile(),
SetConnectedThresholdQuantile() and SetMinimumObjectSize() methods.
SegmentationFilterType::Pointer segmentation = SegmentationFilterType::New();
segmentation->SetReferenceImage(reader->GetOutput()); segmentation->SetBrighterDetails(pyramid->GetSupFilter());
segmentation->SetDarkerDetails(pyramid->GetInfFilter()); segmentation->SetSeedsQuantile(seedsQuantile);
segmentation->SetConnectedThresholdQuantile(segmentationQuantile);
segmentation->SetMinimumObjectSize(minObjectSize);
The pipeline is executed bu calling the Update() method.
Finally, we get an iterator to the list generated as output for the segmentation and we use it to iterate through
the list and write the images to files.
OutputListIteratorType it = segmentation->GetOutput()->Begin(); WriterType::Pointer writer;
int index = 1; std::stringstream oss; while (it != segmentation->GetOutput()->End())
{ oss << outputFilenamePrefix << index << "." << outputFilenameSuffix; writer = WriterType::New();
writer->SetInput(it.Get()); writer->SetFileName(oss.str().c_str()); writer->Update();
std::cout << oss.str() << " file written." << std::endl; oss.str(""); ++index; ++it; }
The user will pay attention to the fact that the list contains first the brighter details segmentation from
higher scale to lower, and then the darker details in the same order.